Bacterial diversity in goat milk with clinical mastitis in Ecuador
Main Article Content
Abstract
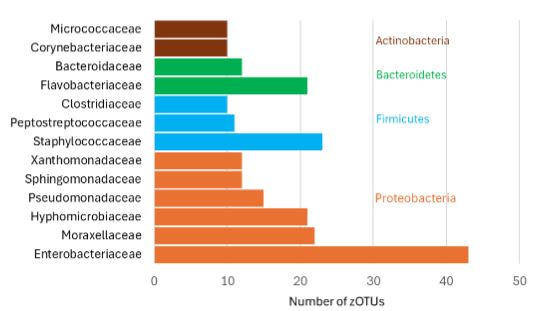
Mastitis is an inflammation of the mammary gland that significantly affects goat milk production and quality, leading to economic losses for Ecuadorian farmers. This study aimed to research the bacterial diversity associated with clinical mastitis in goats from Ecuador. Milk samples were collected from goats showing macroscopic evidence of clinical mastitis for subsequent DNA extraction. The 16S rRNA gene was amplified and sequenced using Illumina MiSeq technology for metagenomic analysis. Sequences were quality filtered and clustered into Zero-radius Operational Taxonomic Units (zOTUs). They were then taxonomically grouped to classify the bacterial species present in the milk. The analysis revealed a high diversity of bacterial communities, with 550 zOTUs belonging to 12 phyla. Proteobacteria and Firmicutes emerged as the predominant phyla, harboring diverse families which include: Enterobacteriaceae (Proteobacteria) and Staphylococcaceae (Firmicutes). Notably, a significant portion (86.91%) of the zOTUs identified belonged to families with unknown functionalities related to mastitis. These findings offer an initial and valuable overview of the bacterial diversity associated with clinical mastitis in goats, as well as highlighting the potential presence of uncharacterized mastitis-related bacteria that could be relevant for this disease. Future studies focusing on species level identification and functional characterization are needed to develop strategies for the prevention and control of mastitis in Ecuadorian goat production.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
National Center for Animal and Plant Health (CENSA)References
FAO. (2020) Dairy Market Review - Overview of Global Dairy Market Developments in 2019.
Prosser CG. Navigating Food Analysis: Practical Tips and Tricks from Sample Prep to LC. J. Food Sci. 2021; 86: 257-265. https:// doi.org/10.1111/1750-3841.15574].
ALKaisy QH, Al-Saadi J, Al-Rikabi AK, Altemimi AB, Hesarinejad A, Abeledmaksoud TG. Exploring the health benefits and functional properties of goat milk proteins. Food Sci Nutr. 2023; 11(10): 5641-5656. doi: 10.1002/fsn3.3531.
Mirzaei H, Chaleshtori RS. Role of fermented goat milk as a nutritional product to improve anemia, J Food Biochem. 2021; 46, e13969. https://doi.org/ 10.1111/jfbc.13969
Ajmone-Marsan P, Colli L, Han JL, Achilli A, Lancioni H, Joost S, Crepaldi P, Pilla F, Stella A, Taberlet P, Boettcher P, Negrini R, Lenstra JA. The characterization of goat genetic diversity: Towards a genomic approach. Small Rumin Res. 2014; 121(1):58-72. doi:10.1016/j.smallrumres.2014.06.010.
Ginja C, Cortés O, Gama LT, Delgado JV, Amills M, De Sousa C, et al. (2018) Conservation of Goat Populations from Southwestern Europe Based on Molecular Diversity Criteria. In: Simões J, Gutiérrez C, eds. Sustainable Goat Production in Adverse Environments: Volume I. Springer, Cham pp 509-5335. doi:10.1038/ismej.2011.41.
Nguyen VD, Nguyen CO, Chau TML, Nguyen DQD, Han AT, Le TTH. Goat Production, Supply Chains, Challenges, and Opportunities for Development in Vietnam: A Review. Animals. 2023;13(15):2546. doi: 10.3390/ani13152546
Polveiro RC, Vidigal PM, Mendes TA, Yamatogi RS, Lima MC, Moreira MA. Effects of enrofloxacin treatment on the bacterial microbiota of milk from goats with persistent mastitis. Sci Rep. 2020; 10(1):4421. doi:10.1038/s41598-020-61407-2.
Oikonomou G, Bicalho ML, Meira E, Rossi RE, Foditch C, Machado VS, Teixeira AG, Santisteban C, Schukken YH, Bicalho RC. Microbiota of Cow’s Milk; Distinguishing Healthy, Sub-Clinically and Clinically Diseased Quarters. Guan LL, ed. PLoS ONE. 2014; 9(1): e85904. doi:10.1371/journal.pone.0085904.
Azooz MF, El-Wakeel SA, Yousef HM. Financial and economic analyses of the impact of cattle mastitis on the profitability of Egyptian dairy farms. Vet World. 2020; 13(9):1750-1759. doi: 10.14202/vetworld.2020.1750-1759.
Podhorecká K, Borková M, Šulc M, Šulc M, Seydlová R, Hedvika Dragounová, H, Švejcarová M, Peroutková J, Elich O. Somatic Cell Count in Goat Milk: An Indirect Quality Indicator. Foods. 2021; 10(5), 1046; https://doi.org/10.3390/foods10051046.
Menzies P. Udder Health for Dairy Goats. Veterinary Clinics: Food Animal Practice. 2021; 37:1, 149 - 174
Novac CS, Andrei S. The Impact of Mastitis on the Biochemical Parameters, Oxidative and Nitrosative Stress Markers in Goat’s Milk: A Review. Pathogens. 2020; 9(11):882. doi:10.3390/pathogens9110882.
Rahimi H, Tukmechi A, Rashidian E. Genetic diversity of Brucella melitensis isolates from sheep and goat milk in Iran. Veterinary Research Forum. 2023; 14 (12): 649-657 doi: 10.30466/vrf.2023.1988859.3768
Contreras A, Sierra D, Sánchez A, Corrales JC, Marco JC, Paape MJ, Gonzalo C. Mastitis in small ruminants. Small Rumin Res. 2007; 68(1-2):145-153. doi:10.1016/j.smallrumres.2006.09.011.
Polveiro RC, Vidigal PMP, De Oliveira Mendes TA, Yamatogi RS, Da SilvaLS, Fujikura JM, et al. Distinguishing the milk microbiota of healthy goats and goats diagnosed with subclinical mastitis, clinical mastitis, and gangrenous mastitis. Front Microbiol. 2022; 13:918706. doi:10.3389/fmicb.2022.918706.
Deddefo A, Mamo G, Leta S, Amenu K. Prevalence and molecular characteristics of Sthaphylococcus aureus in raw milk and milk products in Ethiopia: a systematic review and meta-analisis. Int. J. Food Contam. 2022; 9:8 https://doi.org/ 10.1186/s40550-022-00094-5
Nagao PE, BurkovskiA, Mattos-Guaraldi AL. Editorial: Streptococcus spp. and Corynebacterium spp.: Clinical and Zoonotic Epidemiology, Virulence Potential, Antimicrobial Resistance, and Genomic Trends and Approaches. Front Microbiol. 2022; 28 (13):867210. doi: 10.3389/fmicb.2022.867210
McInnisEA, Kalanetra KM, Mills DA, Maga EA. Analysis of raw goat milk microbiota: Impact of stage of lactation and lysozyme on microbial diversity. Food Microbiol. 2015; 46:121-131. doi:10.1016/j.fm.2014.07.021.
Zhang F, Wang Z, Lei F, Wang Z, Jiang S, Peng Q, Zhang J, Shao Y. Bacterial diversity in goat milk from the Guanzhong area of China. J Dairy Sci. 2017; 100 (10):7812-7824. doi:10.3168/jds.2017-13244.
Ngara TR, Zhang H. Recent Advances in Function-Based Metagenomic Screening. Genomics Proteomics Bioinformatics. 2018; 16 (6):405-415. doi:10.1016/j.gpb.2018.01.002.
Ahmad T, Singh RS, Gupta G, Sharma A, Kaur B. Metagenomics in the Search for Industrial Enzymes. In: Advances in Enzyme Technology. Elsevier. 2019; 419-451.
Wright RJ, Gibson MI, Christie-Oleza JA. Understanding microbial community dynamics to improve optimal microbiome selection. Microbiome. 2019; 7(1):85. doi:10.1186/s40168-019-0702-x.
Win Epi (Working in epidemiology) 2024. [Citado Sept 29]. Disponible en: http:// www.winepi.net/sp/index.htm
ESPAC (Instituto Ecuatoriano de Estadísticas y Censos) 2023 [Citado Sept 29]. Disponible en: https://www.ecuadorencifras.gob.ec/estadisticasagropecuarias-2/
Herlemann, DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011; 5(10):1571-1579. doi:10.1038/ismej.2011.41.
Andrés S. FastQC: A Quality Control tool for High Throughput Sequence Data. 2012. https:// www.bioinformatics.babraham.ac.uk/projects/fastqc/
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30 (15):2114-2120. doi:10.1093/bioinformatics/btu170.
Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013; 10(10):996-998.
Edgar RC. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Valencia A, ed. Bioinformatics. 2018; 34(14):2371-2375. doi:10.1038/nmeth.2604.
Herrera P, Suárez JP, Sánchez-Rodríguez, A, Molina MC, Prieto M, Méndez M. Many broadly-shared mycobionts characterize mycorrhizal interactions of two coexisting epiphytic orchids in a high elevation tropical forest. Fungal Ecol. 2019; 39:26-36. doi:10.1016/j.funeco.2018.11.003.
Edgar RC. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv, 2016. doi:10.1093/bioinformatics/bty113.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon ta. Nat Methods. 2016; 13(7):581-583. doi:10.1038/nmeth.3869.
SoberónM J, Llorente BJ. The Use of Species Accumulation Functions for the Prediction of Species Richness. Conserv Biol.1993; 7(3):480-488. doi:10.1046/j.1523-1739.1993.07030480.x.
Jiménez-ValverdeA, Horta J Las curvas de acumulación de especies y la necesidad de evaluar la calidad de los inventarios biológicos. Rev Ibérica Aracnol2003; 8:151-161.
Anil,N.S, JanusA., Deepa, P.M. Shyma VH, RajasekharR., Habeeb BB. Anand LF. Clinicopathological findings and etiological characterization in caprine gangrenous mastitis. Small Rumin Res. 2024 241: 107389 https://doi.org/ 10.1016/j.smallrumres.2024.107389
RibeiroMG, Lra GHB, Bicudo SD, Souza AVG, Salerno T, Siqueira AK, GeraldoJS. An unusual gangrenous goat mastitis caused by Staphylococcus aureus, Clostridium perfringens and Escherichia coli co-infection. Arq Bras Med Veterinária E Zootec. 2007; 59(3):810-812. doi:10.1590/S0102-09352007000300037.
Niyazbekova Z, Yao XT, Liu MJ, Bold N, Tong JZ, Chang Jj, Wen Y, Li L, WangY, Chen DK, Ma WT. Compositional and functional comparison of the microbiota in the colostrum and mature Milk of dairy goats. Animals. 2020; 10(11): 1955 https://doi.org/ 10.3390/ani10111955
Quigley L, O’Sullivan O, Stanton C, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. The complex microbiota of raw milk. FEMS Microbiol Rev. 2013; 37(5):664-698. doi:10.1111/1574-6976.12030.
Breitenwieser F, Doll EV, Clavel T, Scherer S, Wenning M. Complementary Use of Cultivation and High-Throughput Amplicon Sequencing Reveals High Biodiversity Within Raw Milk Microbiota. Front Microbiol. 2020; 11:1557. doi:10.3389/fmicb.2020.01557.
Nelli A, Voidarou C, Venardou B, Fotou K, Tsinas A, Bonos E, Fthenakis GC, Skoufos I, Tzora A. Antimicrobial and Methicillin Resistance Pattern of Potential Mastitis-Inducing Staphylococcus aureus and Coagulase-Negative Staphylococci Isolates from the Mammary Secretion of Dairy Goats. Biology. 2022; 11(11):1591. doi:10.3390/biology11111591.
Jabbar A, Saleem MH, Iqbal MZ, Qasim M, Ashraf M, Tolba MM, et al. Epidemiology and antibiogram of common mastitis-causing bacteria in Beetal goats. Vet World. 2020; 13(12):2596-2607. doi:10.14202/vetworld.2020.2596-2607.
Pereira CS, Santos LM, Machado LS, Melo DA, Coelho S, Pereira V, Souza M, Nascimiento E. Proteomics characterization of Staphylococcus spp. from goat mastitis and phenogeno-typical assessment of resistance to beta-lactamics. Pesqui Veterinária Bras. 2021; 41:e06129. doi:10.1590/1678-5150pvb-6129.
Hoving-Bolink RA, Antonis AF, te Pas MFW, Schokker D. An observational study of the presence and variability of the microbiota composition of goat herd milk related to mainstream and artisanal farm management. PLOS ONE. 2023; 18(10): e0292650. https://doi.org/10.1371/journal.pone.0292650.
Tormo H, Delacroix-A, Lopez C, Lekhal DA, Roques C. Farm Management Practices and Diversity of the Dominant Bacterial Species in Raw Goat’s Milk. Int J Dairy Sci. 2010; 6(1):29-43. doi:10.3923/ijds.2011.29.43.
Barraza A, Montes-Sánchez JJ, Caamal-Chan MG, Loera-Muro A. Characterization of microbial communities from rumen and large intestine of lactating creole goats grazing in arid plant communities. Microbiology. 2021;167(10). doi:10.1099/mic.0.001092.
Neviani E, Bottari B, Lazzi C, Gatti M. New developments in the study of the microbiota of raw-milk, long-ripened cheeses by molecular methods: the case of Grana Padano and Parmigiano Reggiano. Front Microbiol. 2013; 4 (36): 1-14. doi:10.3389/fmicb.2013.00036.
Yang Z, Ni L, Tian W, Chi H. Screening and Identification of Goat-Milk-Derived Lactic Acid Bacteria with Bacteriocin-like Activity and Probiotic Potentials. Microorganisms. 2023-, 11(4):849. doi:10.3390/microorganisms11040849.
Toquet M, Gómez-Martín betaller E. Review of the bacterial composition of healthy milk, mastitis milk and colostrum in small ruminants. Research in Veterinary Science. 2021; 140 1-5 https://doi.org/ 10.1016/j.rvsc.2021.07.022.