SYBR Green-based real-time PCR assay for the detection of porcine epidemic diarrhea virus
Main Article Content
Abstract
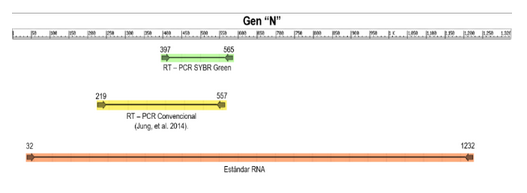
Porcine epidemic diarrhea (PED) is a contagious disease caused by its virus, classified within the genus Alphacoronavirus, family Coronaviridae. This virus causes diarrhea, vomiting and weight loss in pigs of all ages, especially in neonatal piglets, with morbidity and mortality of up to 100%. In Ecuador, the disease was diagnosed for the first time in 2014 by conventional or endpoint PCR. Currently, there is a need for an ultrasensitive and specific assay, which allows working a large number of samples to facilitate epidemiological surveillance of new outbreaks and detect carrier animals. A real-time PCR assay intercalating DNA dyes such as SYBR Green was developed and assessed to detect PED virus (PEDv) in swine feces with primers targeting the N-gene of the virus. A PEDv RNA standard synthesized in the laboratory was used in the assessment of the analytical sensitivity. The detection limit of the developed assay was determined to be 4 copies of PEDv RNA, compared to endpoint PCR which was 100 times more sensitive. Assay specificity was based on the possibility of detecting false positives using the melting curve. Cycle threshold (Ct) values in intra- and inter-assay analysis showed valid coefficients of variation. When assessing diagnostic performance, the assay detected 18 positive samples out of a total of 52 for 100 % agreement with the endpoint PCR. It is concluded that the developed SYBR Green-based real-time PCR assay is highly sensitive, specific, reproducible and rapid for the detection of PEDv in swine intestinal mucosa homogenates.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
National Center for Animal and Plant Health (CENSA)References
Liu Q, Gerdts V. Transmissible gastroenteritis virus of pigs and porcine epidemic diarrhea virus (coronaviridae). Encyclopedia of Virology. Elsevier; 2021. p. 850-3.
Jung K, Saif LJ. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet J. 2015;204(2):134-43.
Wang L, Byrum B, Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerging Infect Dis. 2014;20(5):917-9.
Song D, Moon H, Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res. 2015;4(2):166-76.
Garrido A, Barrera M, Vaca M, Acosta A. Primer reporte de diagnóstico molecular de diarrea epidémica porcina en Ecuador. Ecuador es Calidad. 2015;2(2): 15-19.
Barrera M, Garrido-Haro A, Vaca MS, Granda D, Acosta-Batallas A, Pérez LJ. Tracking the origin and deciphering the phylogenetic relationship of porcine epidemic diarrhea virus in Euador. Biomed Res Int. 2017 ;2017:2978718. https://doi.org/10.1155/2017/2978718)
Olech M. Current state of molecular and serological methods for detection of porcine epidemic diarrhea virus. Pathogens. 2022;11(10): 1074 https://doi.org/10.3390/pathogens11101074.
Jung K, Saif LJ, Wang Q. Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020;286:198045):1-13
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. Epub 2012 Jun 22. PMID: 22730293; PMCID: PMC3424584.)
Jung K, Wang Q, Scheuer KA, Lu Z, Zhang Y, Saif LJ. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerging Infect Dis. 2014;20(4):662-5.
Vaerman JL, Saussoy P, Ingargiola I. Evaluation of real-time PCR data. J Biol Regul Homeost Agents. 2004;18(2):212-4.
Vlasova AN, Marthaler D, Wang Q, Culhane MR, Rossow KD, Rovira A, et al. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerging Infect Dis. 2014;20(10):1620-8.
Li F, Zeng Y, Zhang R, Peng K, Jiang C, Xu Z, et al. Genetic variations in S gene of porcine epidemic diarrhoea virus from 2018 in Sichuan Province, China. Vet Med Sci. 2020;6(4):910-8.
Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151-78.
Siddell S, Wege H, ter Meulen V. The structure and replication of coronaviruses. Curr Top Microbiol Immunol. 1982;99:131-63.
Huang Y-W, Dickerman AW, Piñeyro P, Li L, Fang L, Kiehne R, et al. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013;4(5):e00737-13. doi:10.1128/mBio.00737-13.
Zhou X, Zhang T, Song D, Huang T, Peng Q, Chen Y, et al. Comparison and evaluation of conventional RT-PCR, SYBR green I and TaqMan real-time RT-PCR assays for the detection of porcine epidemic diarrhea virus. Mol Cell Probes. 2017;33:36-41.
Han H-Y, Zheng H-H, Zhao Y, Tian R-B, Xu P-L, Hou H-L, et al. Development of a SYBR green I-based duplex real-time fluorescence quantitative PCR assay for the simultaneous detection of porcine epidemic diarrhea virus and porcine circovirus 3. Mol Cell Probes. 2019;44:44-50.
Zheng L-L, Cui J-T, Han H-Y, Hou H-L, Wang L, Liu F, et al. Development of a duplex SYBR Green? based real-time PCR assay for detection of porcine epidemic diarrhea virus and porcine bocavirus3/4/5. Mol Cell Probes. 2020;51:101544.
Lekanne Deprez RH, Fijnvandraat AC, Ruijter JM, Moorman AFM. Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR green I depend on cDNA synthesis conditions. Anal Biochem. 2002;307(1):63-9.
Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245(2):154-60.
Duan C. An updated review of porcine deltacoronavirus in terms of prevalence, pathogenicity, pathogenesis and antiviral strategy. Front Vet Sci. 2021;8:811187.
Zhou P, Fan H, Lan T, Yang X-L, Shi W-F, Zhang W, et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018 ;556(7700):255-8.
Bolivar AM, Rojas A, Lugo PG. PCR y PCR-Múltiple: parámetros críticos y protocolo de estandarización. Avances en Biomedicina. 20143(1): 25-33
World Organisation for Animal Health. Chapter 2.1.2. Biotechnology Advances in the Diagnosis of Infectious Diseases. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals [Internet]. 2022 [cited 2023 Jun 9]. p. 20. Available from: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/2.01.02_BIOTECH_DIAG_INF_DIS.pdf.