Genetic variability characterization of rice pathogenic strains of Pseudomonas spp.
Main Article Content
Abstract
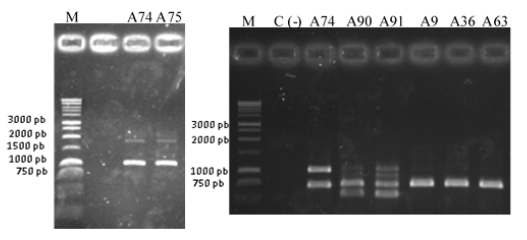
The objective of this work was to characterize the genetic variability of strains of Pseudomonas spp. that cause Sheath Brown Rot of rice in Cuba, through the analysis of genomic regions. A first grouping was performed based on the genetic fingerprint analysis by PCR, and then the analysis was developed through rep-PCR (ERIC, BOX and REP-PCR). In the two types of tests, the strains showed molecular variability, which was evidenced by the formation of three groups of Pseudomonas that circulate in the country. One of them grouped the strains A74 and A75, the second group included the strains A90 and A91, and the last group related most of the strains (A9, A36, A63, PR1 and PR3), the latter identified as Pseudomonas sp. or Pseudomonas fluorescens.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Aquellos autores/as que tengan publicaciones con esta revista, aceptan los términos siguientes:
- Los autores/as conservarán sus derechos de autor y garantizarán a la revista el derecho de primera publicación de su obra, el cual estará simultáneamente sujeto a la Licencia Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) que permite a terceros compartir la obra, siempre que se indique su autor y la primera publicación en esta revista. Bajo esta licencia el autor será libre de:
- Compartir — copiar y redistribuir el material en cualquier medio o formato
- Adaptar — remezclar, transformar y crear a partir del material
- El licenciador no puede revocar estas libertades mientras cumpla con los términos de la licencia
Bajo las siguientes condiciones:
- Reconocimiento — Debe reconocer adecuadamente la autoría, proporcionar un enlace a la licencia e indicar si se han realizado cambios. Puede hacerlo de cualquier manera razonable, pero no de una manera que sugiera que tiene el apoyo del licenciador o lo recibe por el uso que hace.
- NoComercial — No puede utilizar el material para una finalidad comercial.
- No hay restricciones adicionales — No puede aplicar términos legales o medidas tecnológicas que legalmente restrinjan realizar aquello que la licencia permite.
- Los autores/as podrán adoptar otros acuerdos de licencia no exclusiva de distribución de la versión de la obra publicada (p. ej.: depositarla en un archivo telemático institucional o publicarla en un volumen monográfico) siempre que se indique la publicación inicial en esta revista.
- Se permite y recomienda a los autores/as difundir su obra a través de Internet (p. ej.: en archivos telemáticos institucionales o en su página web) antes y durante el proceso de envío, lo cual puede producir intercambios interesantes y aumentar las citas de la obra publicada. (Véase El efecto del acceso abierto).
References
Martínez S, Bao L, Escalante F. Manual de Identificación de Enfermedades y Plagas en el Cultivo del Arroz. Instituto Nacional de Investigación Agropecuaria Uruguay. Boletín de divulgación. 2018;116:66 p.
Pérez HI, Rodríguez I, García RM. Principales enfermedades que afectan al cultivo del arroz en Ecuador y alternativas para su control. Revista Científica Agroecosistemas. 2018; 6(1):16-27.
Pedraza LA, Bautista J, Uribe-Vélez D. Seed-borne Burkholderia glumae infects rice seedling and maintains bacterial population during vegetative and reproductive growth stage. Plant Pathol. J. 2018; 34(5):393-402.
CABI. Pseudomonas syringae pv. oryzae (halo blight). Invasive Species Compendium. 2019; Wallingford, UK: CAB International. https://www.cabi.org/isc/datasheet/44981
Musonerimana S, Bez C, Licastro D, Habarugira G, Bigirimana J, Venturi V. Pathobiomes Revealed that Pseudomonas fuscovaginae and Sarocladium oryzae Are Independently Associated with Rice Sheath Rot. Microbial Ecology. 2020; https://doi.org/10.1007/s00248-020-01529-2
Alam K, Islam M, Li C, Sultana S, Zhong L, Shen Q, et al. Genome Mining of Pseudomonas Species; Diversity and Evolution of Metabolic and Biosynthetic Potential. Molecules. 2021; 26:7524. https://doi.org/10.3390/ molecules26247524
Yi B, Dalpke AH. Revisiting the intrageneric structure of the genus Pseudomonas with complete whole genome sequence information: Insights into diversity and pathogen-related genetic determinants. Infections, Genetics and Evolution. 2022; 97. https://doi.org/10.1016/j.meegid.2021.105183
Peix A, Ramírez ME, Velázquez BE. The current status of Pseudomonas revisited: An update. Infection, Genetics and Evolution. 2018; 57:106-116.
He R, Liu P, Jia B, Xue S, Wang X, Hu J, et al. Genetic diversity of Pseudomonas syringae pv. actinidiae strains from different geographic regions in China. Phytopathology. 2019; 109(3):347-357.
Abdel-Rhman S, Rizk DE. Comparative assessment of different PCR-based typing methods of Pseudomonas aeruginosa isolates. Infection and drug resistance. 2021; 14:1019-1035.
Cottyn B, Regalado E, Lanoot B, De Cleene M, Mew TW, Swings J. Bacterial populations associated with rice seed in the tropical environment. Phytopathology. 2001;91(3):282-92.
Cottyn B, Barrios H, George T, Vera-Cruz CM. Characterization of rice sheath rot from Siniloan, Philippines. Int Rice Res Notes. 2002;27(1):39-40.
ONEI. Oficina Nacional de Estadística e Información. Anuario Estadístico de Cuba 2020. Capítulo 9: Agricultura, ganadería, silvicultura y pesca. Edición 2021. [En línea]. [Consultado: 28 de enero de 2022]. Disponible en: http://www.onei.gob.cu/node/16275
Ravi A, Prathap V, Narayana P, Sokka S, Sivaramakrishnan S. Assessment of Genetic Diversity in Pseudomonas fluorescens using PCR-based Methods. Bioremediation, Biodiversity and Bioavailability. 2011; 5(1):10-16.
Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant molecular biology reporter. 1983; 1(4):19-21.
Onasanya A, Basso A, Somado E, Gasore ER, Nwilene FE, Ingelbrecht I, et al. Development of a combined molecular diagnostic and DNA fingerprinting technique for rice bacteria pathogens in Africa. Biotechnology. 2010; 9(2):89-105.
Sambrook J, Fritsch ER, Maniatis T. Molecular Cloning: A Laboratory Manual (2nd ed.). 1989. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Quibod IL, Grande G, Oreiro EG, Borja FN, Dossa GS, Mauleon R, et al. Rice-Infecting Pseudomonas Genomes Are Highly Accessorized and Harbor Multiple Putative Virulence Mechanisms to Cause Sheath Brown Rot. PloS ONE. 2015; 10(9):e0139256.
CABI. Crop Protection Compendium [CD-Rom]. 2006. Wallingford, Londres: CABI.
Viteri A, Regalado H, Toaza A. Identificación de Pseudomonas fluorescens y Pseudomonas marginalis, causantes del síndrome del peridermo “Pink Eye” en papa. Revista Científica Ecuatoriana. 2020; 7(2):49-59.
Beiki F, Busquets A, Gomila M, Rahimian H, Lalucat J, García E. New Pseudomonas spp. Are Pathogenic to Citrus. PLoS ONE. 2016; 11(2):e0148796.
Aflaha I, Chairul J, Baharuddin A, Kuswinanti T. Molecular identification of bacteria causing grain rot disease on rice. IOP Conference Series: Earth and Environmental Science. 2020;486:012165. doi:10.1088/1755-1315/486/1/012165
Bangratz M, Wonni I, Kini K, Sondo M, Brugidou C, Béna G, et al. Design of a new multiplex PCR assay for rice pathogenic bacteria detection and its application to infer disease incidence and detect co-infection in rice fields in Burkina Faso. PLoS ONE. 2020; 15 (4): e0232115. https://doi.org/10.1371/journal.pone.0232115
Ogier JC, Pages S, Galan M, Barret M, Gaudriault S. rpoB, a promising marker for analizing the diversity of bacterial communities by amplicon sequencing. BMC Microbiology. 2019; 19(171):16 p.
Musonerimana S, Venturi V. The rice sheath rot pathogen Pseudomonas fuscovaginae; microbiome and cell-cell signaling studies. [PhD Thesis in Molecular Biology]. 2020; 156 p. [En línea]. [Consultado: 16 de febrero de 2022]. Disponible en: https://iris.sissa.it/retrieve/handle/20.500.11767/112329/124405/Samson_Thesis_2020.pdf